1- Proteomic analysis by mass spectrometry
PRESENTATION
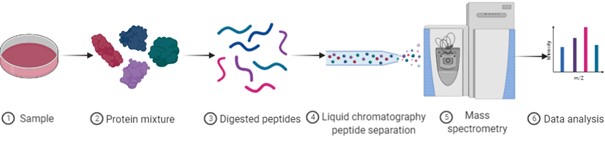
Proteomics allows the identification, characterisation, and quantification of proteins in a biological sample. Proteomic analysis is applicable to a broad range of fields including health, environment, agronomy, etc. Proteomic analysis requires different steps: 1) protein extraction from the biological sample to be studied. 2) protein digestion using enzymes, such as trypsin, to obtain a peptide mixture. 3) Analysis of the peptide mixture by liquid chromatography coupled with a mass spectrometer to obtain peptide fragmentation spectra. All the data are then compared to protein databases using bioinformatics software to identify the proteins present in the sample.
SERVICES
The Mass Spectrometry facility offers the following services as a service or in collaboration:
- Identification and analysis of complex protein mixtures
- Characterisation of post-translational modifications (Phosphorylation, Acetylation, etc.)
- Relative quantification of proteins in complex mixtures (comparison of different conditions) by labelling approaches (iTRAQ/TMT/SILAC) or by label-free approach (LFQ)
- Binding partner discovery after immunoprecipitation
- Structural studies on whole proteins
- Molecular mass measurement of small molecules, peptides, oligonucleotide proteins or synthetic polymers: by MALDI/TOF or ESI/Orbitrap after deconvolution of multicharged ions.
- Development of bioinformatics tools to aid the exploitation of proteomic data
Sample preparation
We handle samples in a wide concentration range (from µg to mg) and from very diverse preparations (gel, liquid, on beads, dry extract).
We take care of the sample processing before analysis by mass spectrometry:
- Enzymatic digestion (different proteases)
- Enrichment step for the characterisation of post-translational modifications
- Isotopic labelling step (TMT, Dimethyl tag) for relative quantification
- Splitting
- Fluorescence peptide assay
- Desalination
Data processing
Analysis software:
Different data analysis software are used for analysing proteomic data:
- Proteome Discoverer 3.1 with Chimerys (Thermo Scientific): DDA data
- Biopharma Finder 3.1 (Thermo Scientific): peptide mapping, whole protein
- Skyline: PRM data
- Spectronaut 18 (Biognosys), DIA-NN: DIA data.
For in-depth and advanced studies, we have available bioinformatic support to develop tools for data visualisation and exploitation. For example, a web tool allows interactive exploration of quantitative proteomic data and enrichment analyses.
Data storage and backup:
All data are stored on our servers for a period of 5 years. After this period, the data are archived.
We provide help for data deposition in public servers (PRIDE), upon request.
Request for analysis
To submit an analysis request, please fill out the MS sample form and bring it with your samples or send it to us by email ( protein.science@ibcp.fr ):
EQUIPMENT
The facility is equipped with 3 mass spectrometers:
| Q Exactive HF Biopharma coupled with a nanoRSLC (Thermo Scientific): nanoelectrospray source, quadrupole-orbitrap analyzers, HCD fragmentation. |  |
|
VOYAGER De Pro (Sciex): MALDI source, TOF analyzer |
 |
|
Exploris 480 (right) coupled with the nanoLC Vanquish NEO (left) (Thermo Scientific): nanoelectrospray source, quadrupole-orbitrap analyzers, HCD fragmentation, FAIMS Pro ion mobility module |


